|
|
3 weeks ago | |
|---|---|---|
| .. | ||
| README.md | 3 weeks ago | |
| assignment.md | 3 weeks ago | |
README.md
Visualizing Distributions
 |
|---|
| Visualizing Distributions - Sketchnote by @nitya |
In the previous lesson, you explored a dataset about the birds of Minnesota. You identified some erroneous data by visualizing outliers and examined differences between bird categories based on their maximum length.
Pre-lecture quiz
Explore the birds dataset
Another way to analyze data is by examining its distribution, or how the data is spread along an axis. For instance, you might want to understand the general distribution of maximum wingspan or maximum body mass for the birds of Minnesota in this dataset.
Let’s uncover some insights about the distributions in this dataset. In your R console, import ggplot2 and the database. Remove the outliers from the database as you did in the previous topic.
library(ggplot2)
birds <- read.csv("../../data/birds.csv",fileEncoding="UTF-8-BOM")
birds_filtered <- subset(birds, MaxWingspan < 500)
head(birds_filtered)
| Name | ScientificName | Category | Order | Family | Genus | ConservationStatus | MinLength | MaxLength | MinBodyMass | MaxBodyMass | MinWingspan | MaxWingspan | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Black-bellied whistling-duck | Dendrocygna autumnalis | Ducks/Geese/Waterfowl | Anseriformes | Anatidae | Dendrocygna | LC | 47 | 56 | 652 | 1020 | 76 | 94 |
| 1 | Fulvous whistling-duck | Dendrocygna bicolor | Ducks/Geese/Waterfowl | Anseriformes | Anatidae | Dendrocygna | LC | 45 | 53 | 712 | 1050 | 85 | 93 |
| 2 | Snow goose | Anser caerulescens | Ducks/Geese/Waterfowl | Anseriformes | Anatidae | Anser | LC | 64 | 79 | 2050 | 4050 | 135 | 165 |
| 3 | Ross's goose | Anser rossii | Ducks/Geese/Waterfowl | Anseriformes | Anatidae | Anser | LC | 57.3 | 64 | 1066 | 1567 | 113 | 116 |
| 4 | Greater white-fronted goose | Anser albifrons | Ducks/Geese/Waterfowl | Anseriformes | Anatidae | Anser | LC | 64 | 81 | 1930 | 3310 | 130 | 165 |
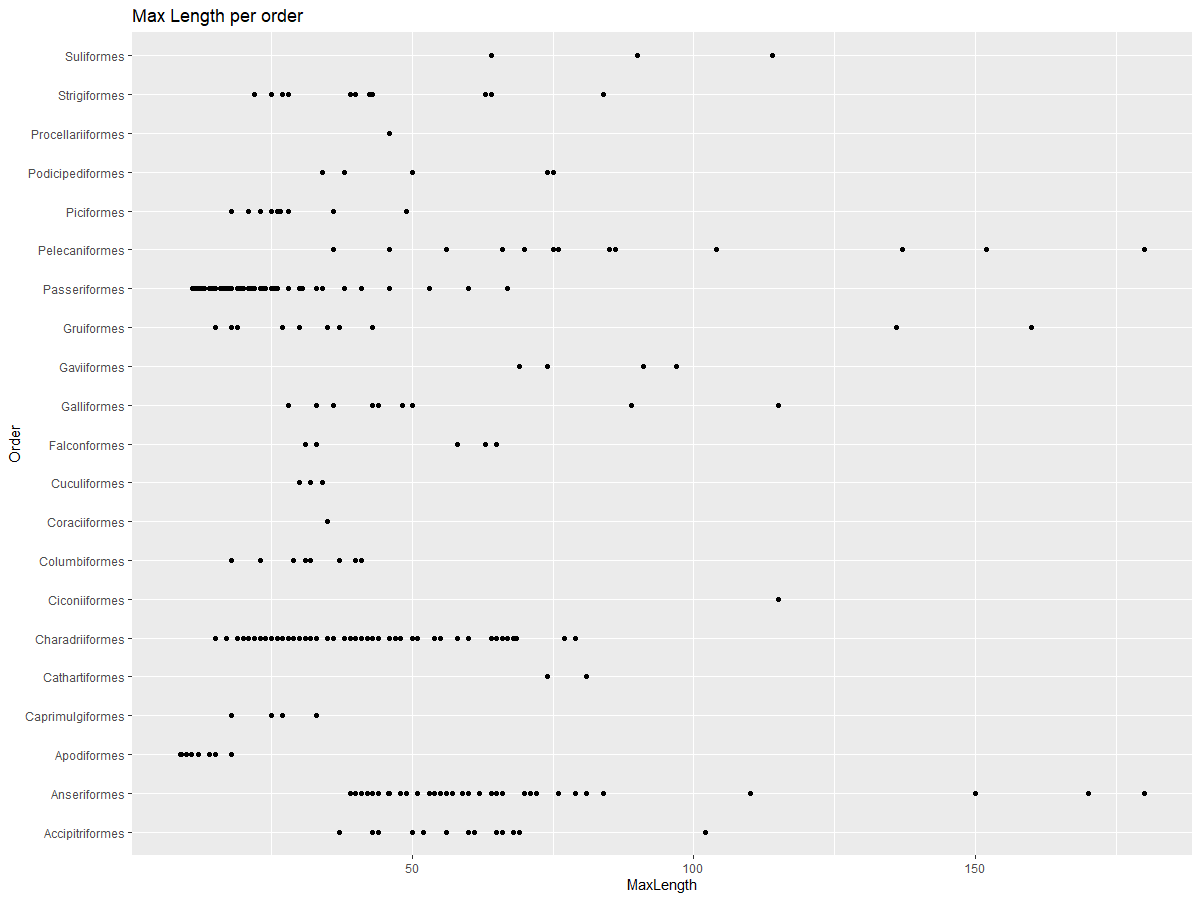
You can quickly visualize how data is distributed by using a scatter plot, as demonstrated in the previous lesson:
ggplot(data=birds_filtered, aes(x=Order, y=MaxLength,group=1)) +
geom_point() +
ggtitle("Max Length per order") + coord_flip()
This provides a general overview of body length distribution per bird Order, but it’s not the best way to display true distributions. For that, histograms are typically used.
Working with histograms
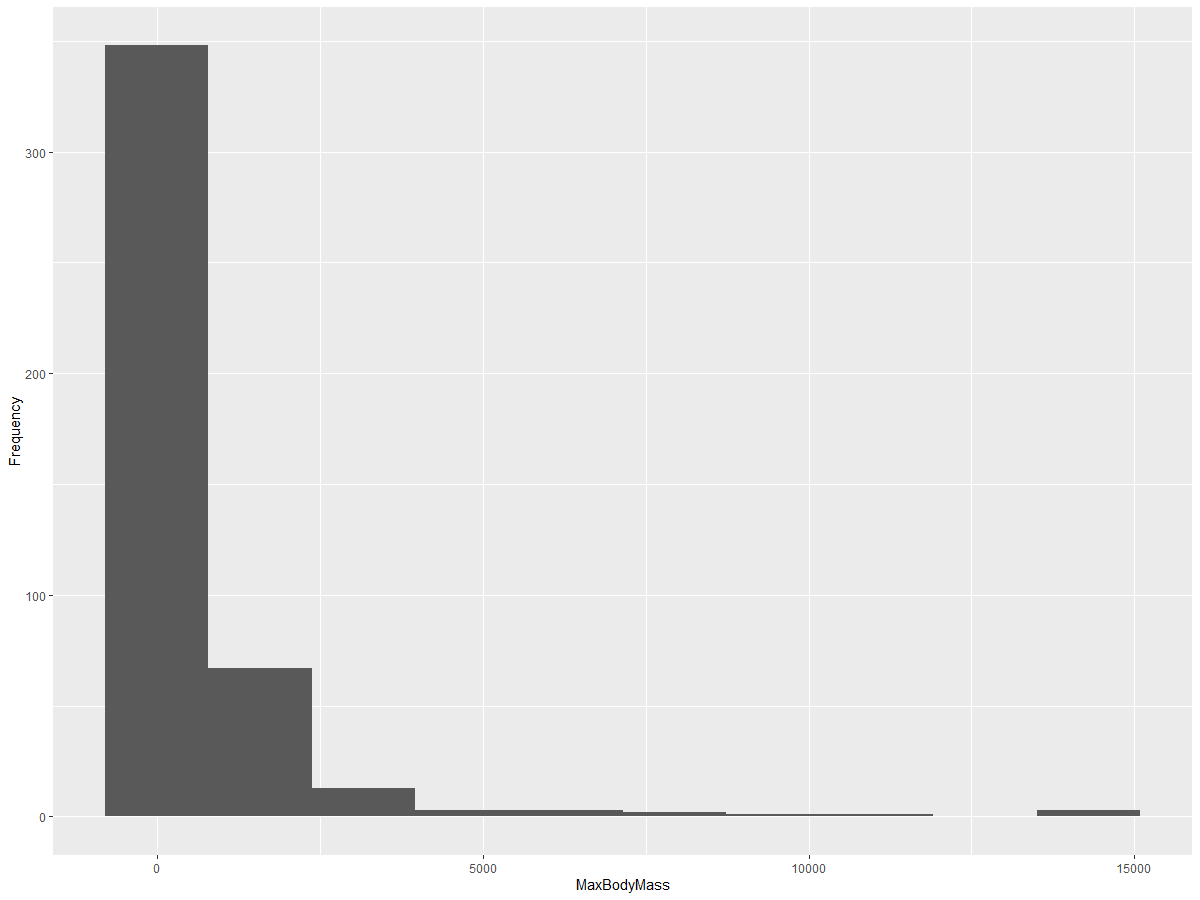
ggplot2 offers excellent tools for visualizing data distribution using histograms. A histogram is similar to a bar chart, but it shows the distribution of data through the rise and fall of the bars. To create a histogram, you need numeric data. You can plot a histogram by specifying the chart type as 'hist'. This chart displays the distribution of MaxBodyMass across the dataset’s numeric range. By dividing the data into smaller bins, it reveals how values are distributed:
ggplot(data = birds_filtered, aes(x = MaxBodyMass)) +
geom_histogram(bins=10)+ylab('Frequency')
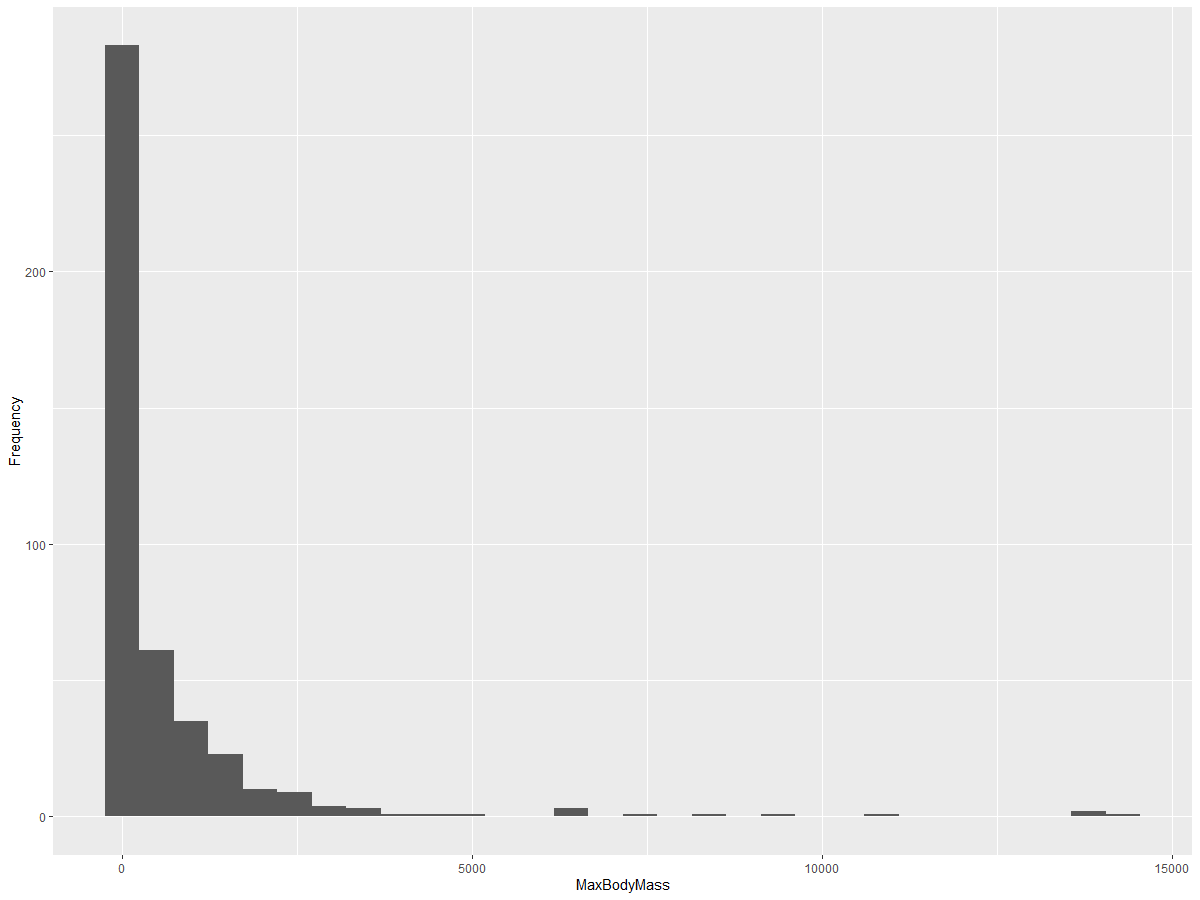
As shown, most of the 400+ birds in this dataset have a Max Body Mass under 2000. You can gain more detailed insights by increasing the bins parameter to a higher value, such as 30:
ggplot(data = birds_filtered, aes(x = MaxBodyMass)) + geom_histogram(bins=30)+ylab('Frequency')
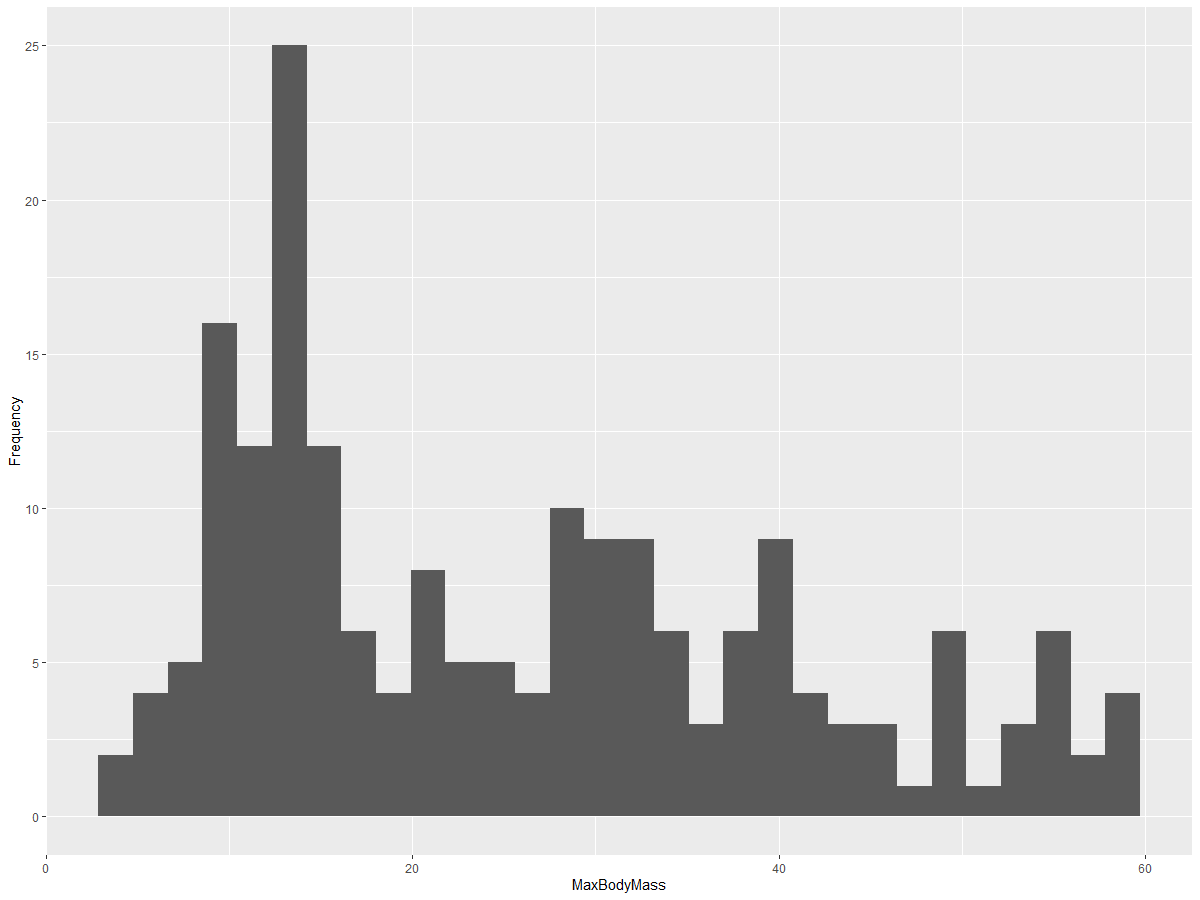
This chart provides a more granular view of the distribution. To create a chart that’s less skewed to the left, you can filter the data to include only birds with a body mass under 60 and display 30 bins:
birds_filtered_1 <- subset(birds_filtered, MaxBodyMass > 1 & MaxBodyMass < 60)
ggplot(data = birds_filtered_1, aes(x = MaxBodyMass)) +
geom_histogram(bins=30)+ylab('Frequency')
✅ Experiment with other filters and data points. To view the full distribution, remove the ['MaxBodyMass'] filter and display labeled distributions.
Histograms also allow for color and labeling enhancements:
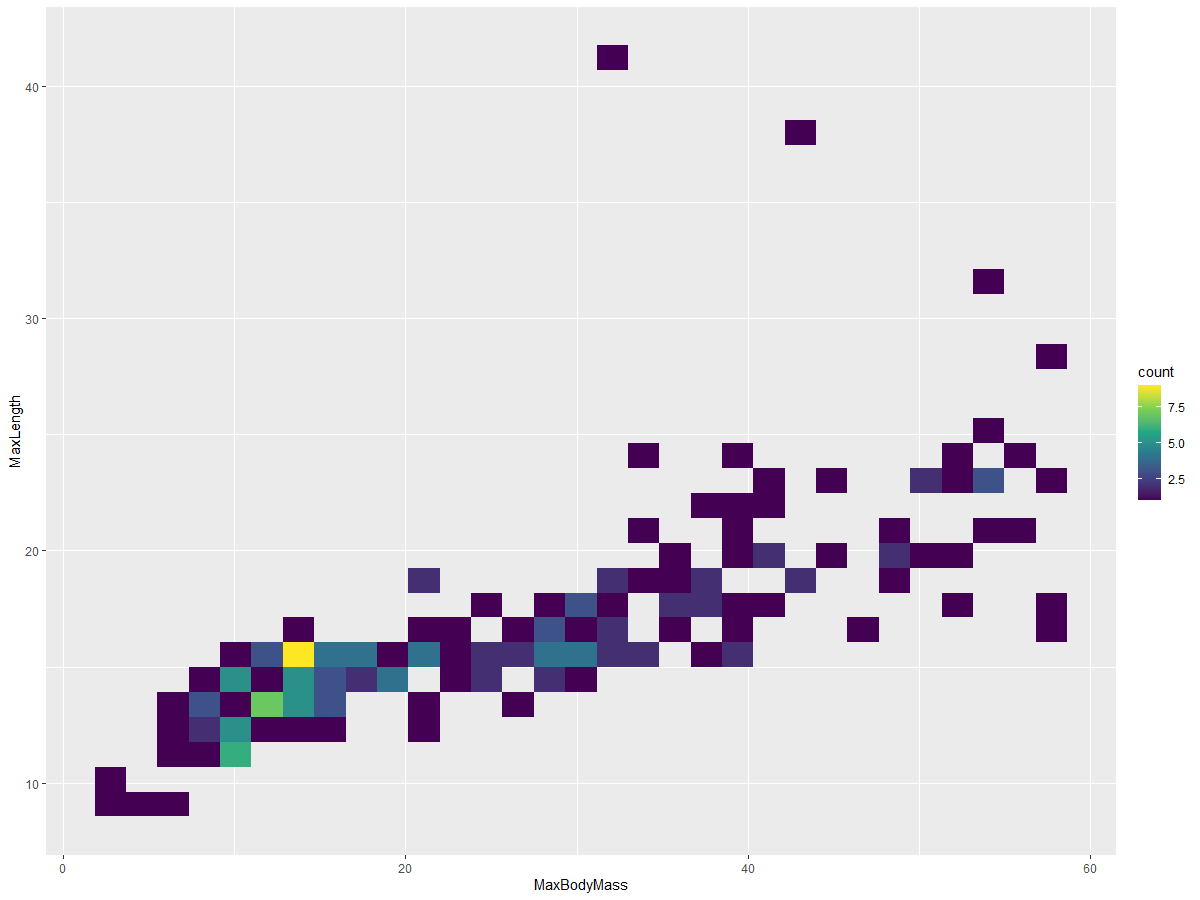
Create a 2D histogram to compare the relationship between two distributions. For example, compare MaxBodyMass vs. MaxLength. ggplot2 provides a built-in method to show convergence using brighter colors:
ggplot(data=birds_filtered_1, aes(x=MaxBodyMass, y=MaxLength) ) +
geom_bin2d() +scale_fill_continuous(type = "viridis")
There seems to be a predictable correlation between these two elements along a specific axis, with one particularly strong point of convergence:
Histograms work well for numeric data by default. But what if you need to analyze distributions based on text data?
Explore the dataset for distributions using text data
This dataset also contains valuable information about bird categories, genus, species, family, and conservation status. Let’s examine the conservation status distribution. What does it look like?
✅ In the dataset, several acronyms are used to describe conservation status. These acronyms are derived from the IUCN Red List Categories, which catalog species' statuses.
- CR: Critically Endangered
- EN: Endangered
- EX: Extinct
- LC: Least Concern
- NT: Near Threatened
- VU: Vulnerable
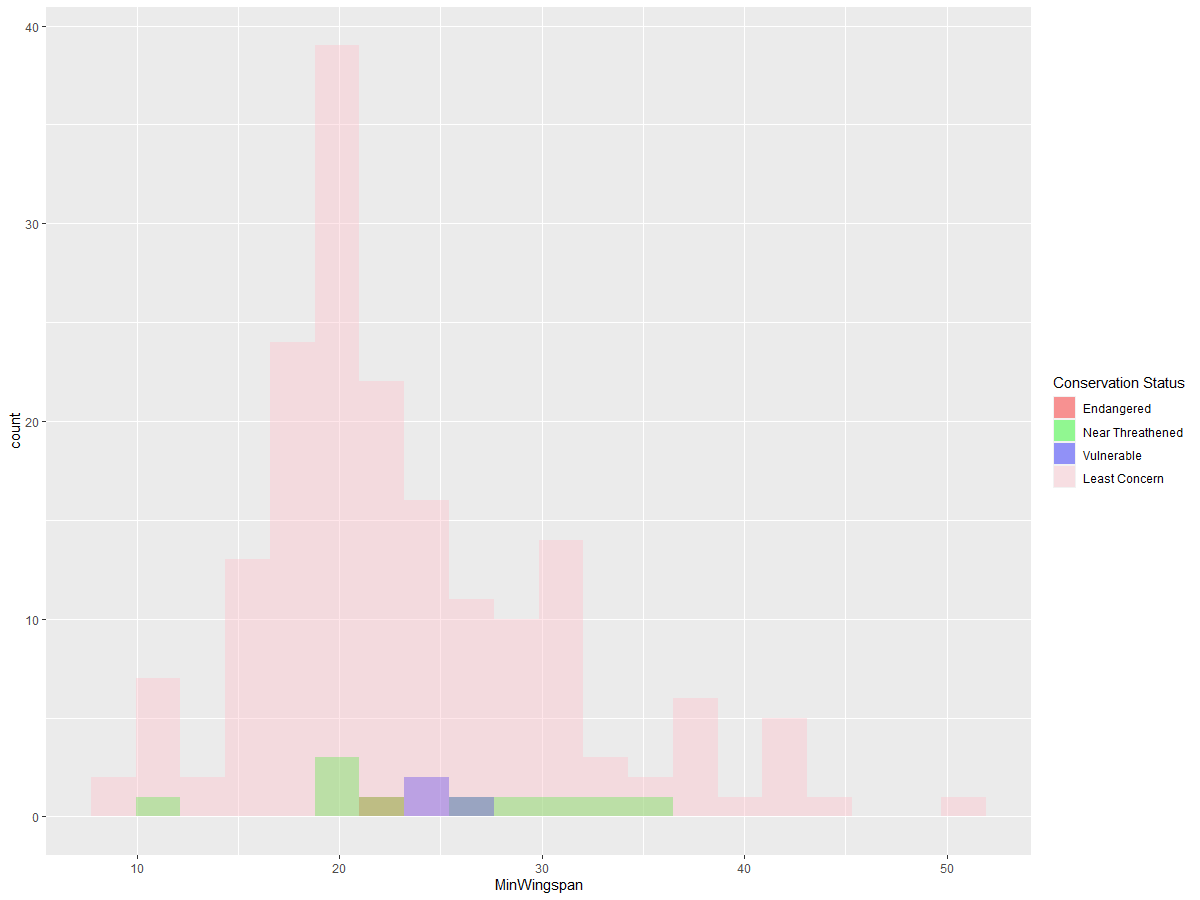
Since these are text-based values, you’ll need to transform the data to create a histogram. Using the filteredBirds dataframe, display conservation status alongside Minimum Wingspan. What do you observe?
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'EX'] <- 'x1'
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'CR'] <- 'x2'
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'EN'] <- 'x3'
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'NT'] <- 'x4'
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'VU'] <- 'x5'
birds_filtered_1$ConservationStatus[birds_filtered_1$ConservationStatus == 'LC'] <- 'x6'
ggplot(data=birds_filtered_1, aes(x = MinWingspan, fill = ConservationStatus)) +
geom_histogram(position = "identity", alpha = 0.4, bins = 20) +
scale_fill_manual(name="Conservation Status",values=c("red","green","blue","pink"),labels=c("Endangered","Near Threathened","Vulnerable","Least Concern"))
There doesn’t appear to be a strong correlation between minimum wingspan and conservation status. Test other elements of the dataset using this method. Try different filters as well. Do you find any correlations?
Density plots
You may have noticed that the histograms we’ve explored so far are ‘stepped’ and don’t flow smoothly in an arc. To create a smoother density chart, you can use a density plot.
Let’s explore density plots now!
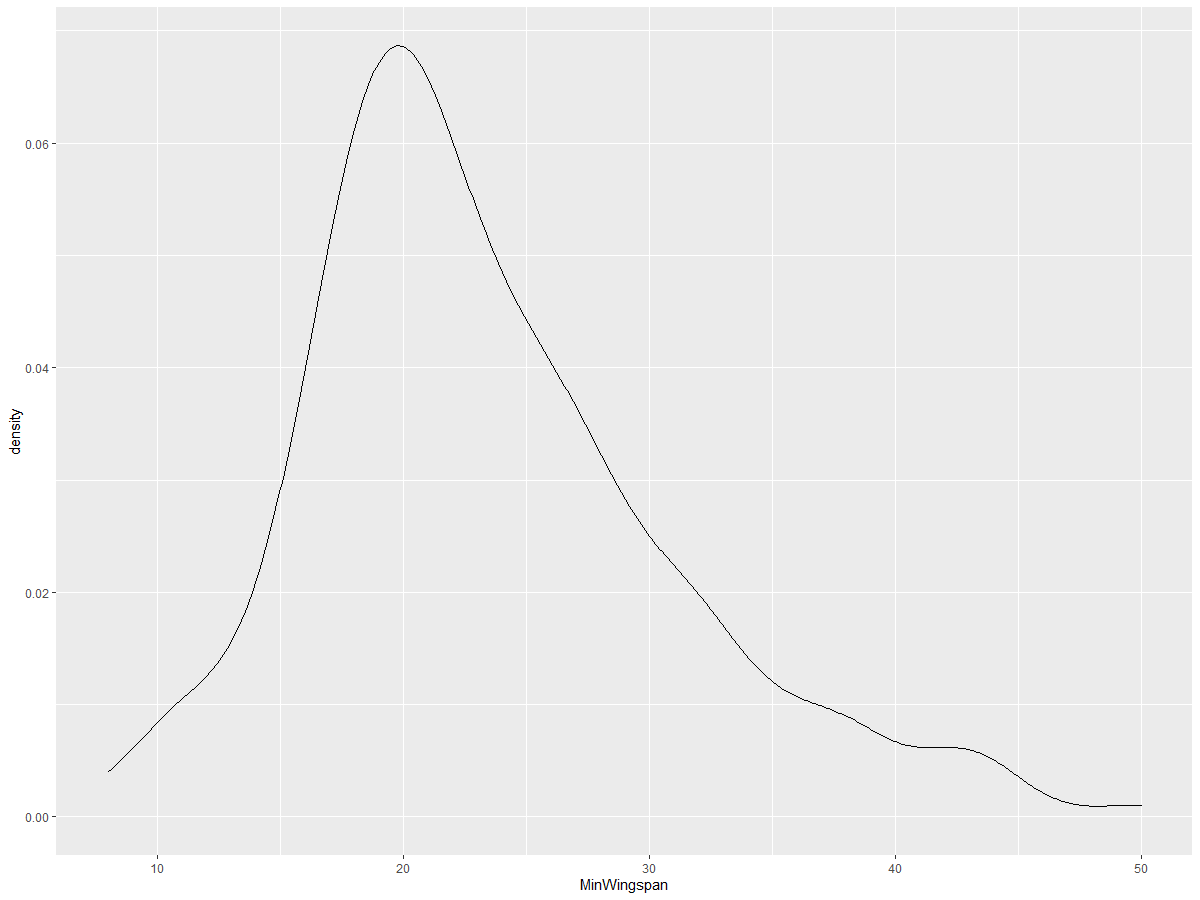
ggplot(data = birds_filtered_1, aes(x = MinWingspan)) +
geom_density()
This plot mirrors the previous one for Minimum Wingspan data but is smoother. If you want to revisit the jagged MaxBodyMass line from the second chart, you can smooth it out using this method:
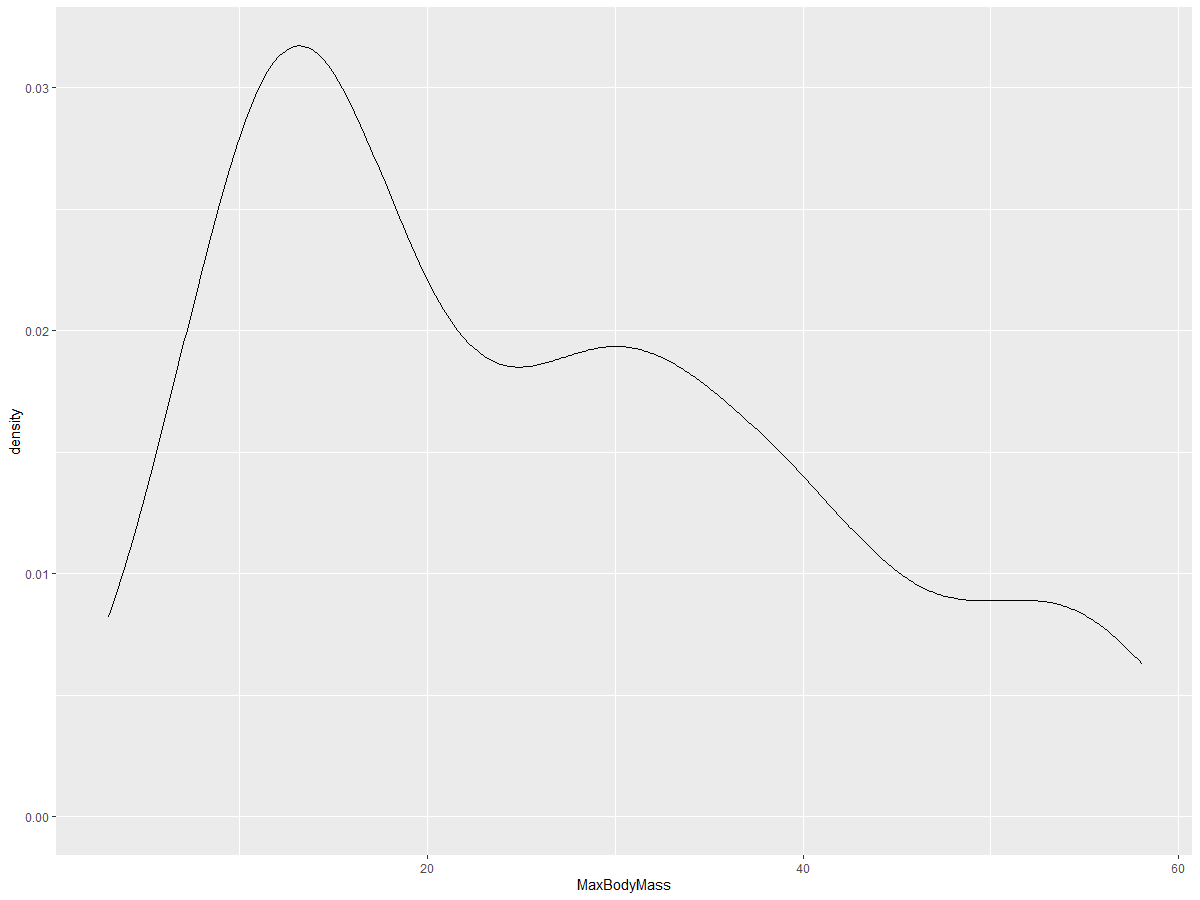
ggplot(data = birds_filtered_1, aes(x = MaxBodyMass)) +
geom_density()
If you prefer a smooth but not overly smooth line, adjust the adjust parameter:
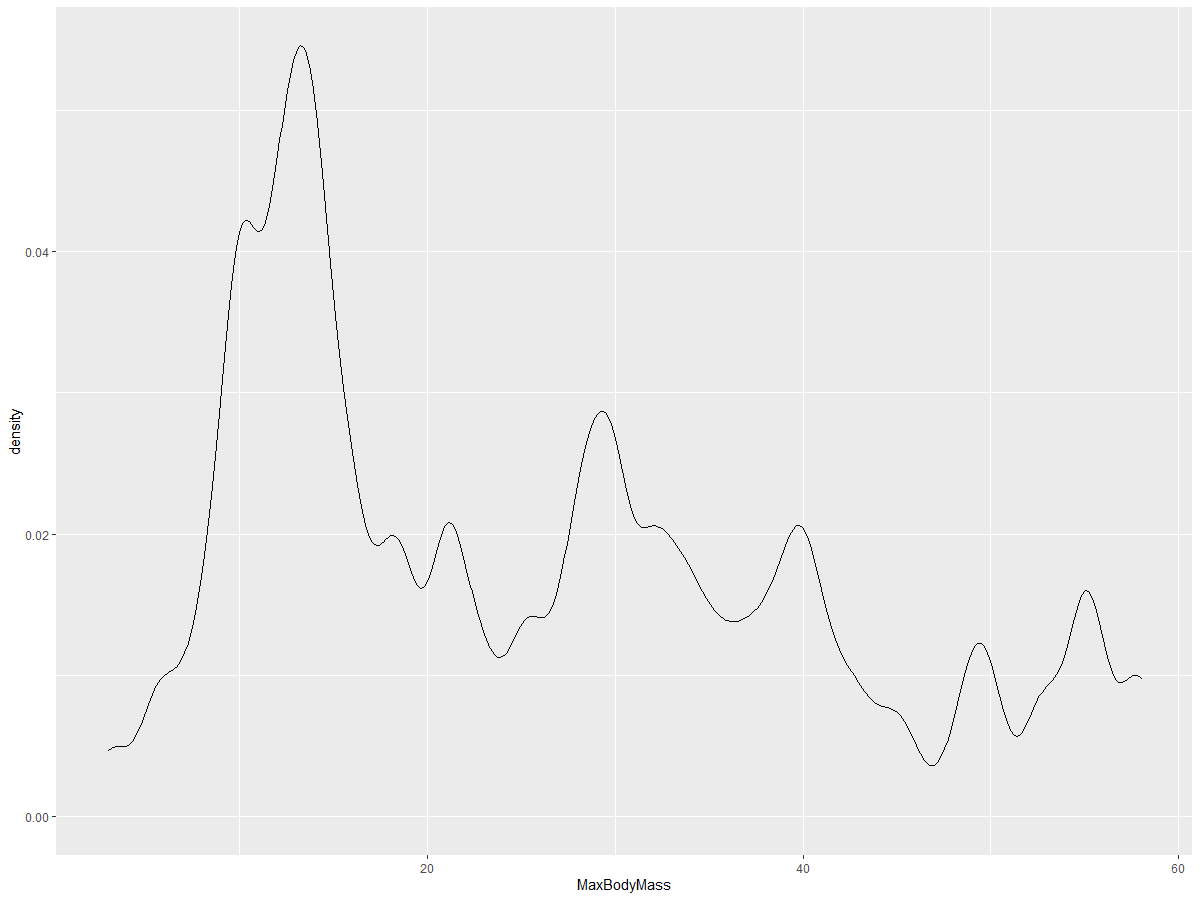
ggplot(data = birds_filtered_1, aes(x = MaxBodyMass)) +
geom_density(adjust = 1/5)
✅ Explore the available parameters for this type of plot and experiment!
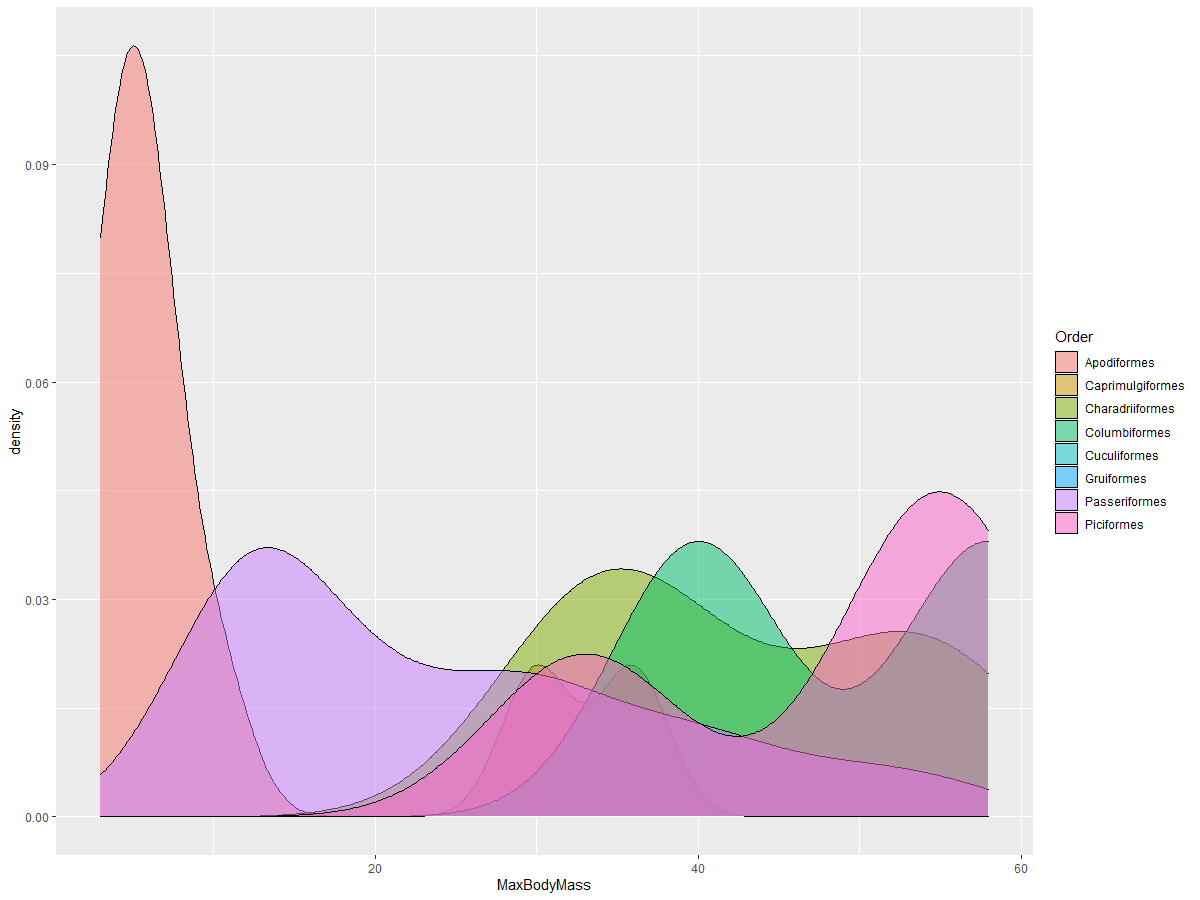
This type of chart provides visually compelling explanations. For instance, with just a few lines of code, you can display the max body mass density per bird Order:
ggplot(data=birds_filtered_1,aes(x = MaxBodyMass, fill = Order)) +
geom_density(alpha=0.5)
🚀 Challenge
Histograms are more advanced than basic scatterplots, bar charts, or line charts. Search online for examples of histograms. How are they used, what do they reveal, and in which fields or areas of study are they commonly applied?
Post-lecture quiz
Review & Self Study
In this lesson, you used ggplot2 to create more advanced charts. Research geom_density_2d(), which generates "continuous probability density curves in one or more dimensions." Read through the documentation to understand its functionality.
Assignment
Disclaimer:
This document has been translated using the AI translation service Co-op Translator. While we aim for accuracy, please note that automated translations may include errors or inaccuracies. The original document in its native language should be regarded as the authoritative source. For critical information, professional human translation is advised. We are not responsible for any misunderstandings or misinterpretations resulting from the use of this translation.